EXECUTIVE OVERVEIW
KNN as a Feature Engine can aid in ensemble learning by quantifying anecdotal knowledge through supervised machine learning.
A good example would be targeting a minority group of customers who are known to have a desirable trait (e.g., similar features/patterns in customer behavior indicative of higher ‘value’ buyers, etc.).
Part two of this two-part series incorporates the KNN Feature Engine from part one into different ensemble models and reviews the findings.
UPDATE 8/8/2023: Full code can be found on Github
THE DETAILS
Table of Contents
- Part One (Updates)
- Code: KNN as a Feature Engine (Part One Updated)
- Code: Testing K (Part One Updated)
- Code: Ensemble Learning (Part Two)
- Part Two
- Final Considerations
- Addendum: Part 1 Updates
Part One (Updates)
Since the last post, there have been momentous changes to the code that warrant a part one update. In this section, we will go over the high-level details (as it pertains to continuing with part two) so that we can continue with our ‘ensemble-journey’.
Summary part one updates:
- The prototype ‘bias’ feature is now working! This allows us to train, fit, and generalize models as the data has real abnormalities to exploit.
- A validate/test/deploy paradigm has been adopted allowing us to test trained models on a hold-out set to authenticate generalization capabilities! This also allows us to control data leakage enabling stronger, more reliant models!
- An ensemble of changes – including new functions, loops, and other coding enhancements- makes the code more resilient, readable, functionable, and robust than before!
As it relates to part one, the training output needs to be updated to show new outputs based on training changes (see below)
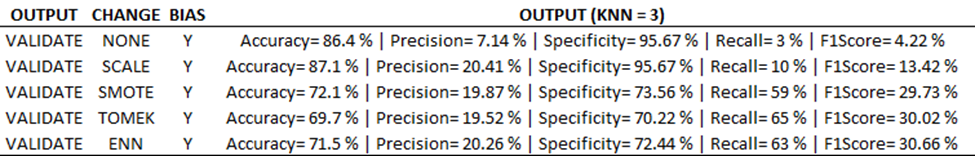
As you can see, we are still tracking with a standard ‘value-increase’ as we process this imbalanced data. However, the surprise comes when applying the ‘best’ model to the validation set. You can see a nearly 20% decrease in precision, recall, F1 score (17.7, 15.3, 17.2% decrease respectively)! This is a huge hit to the most important metrics, and it also portrays a more important story: our model doesn’t generalize to new data well. This means we were overfitting the noise and not the signal.
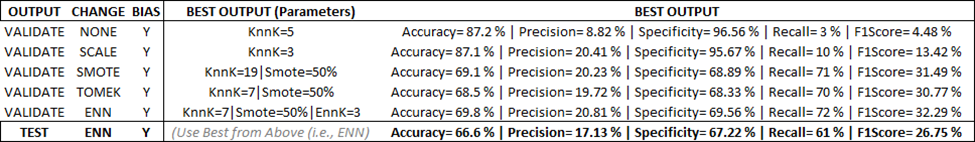
To combat this, we select another model that predicts and generalizes well, and we use that for our ensemble learning model.

Here is a ChatGPT excerpt on ‘considering generalization’:


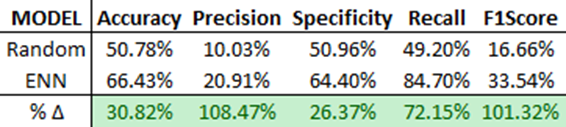
Finally, we deploy this model and compare the results against the ‘randomized control’ benchmark from part one in this series. You can see the massive gains in predictive power over random chance alone. This output will feed into part two of this model where we combine this feature-engine/pseudo-label into our ensemble model.
For more information on the part one update and the training data, please reference the addendum at the bottom of this article.
Code: KNN as a Feature Engine (Part One Updated) (R)
##########################
## Establish Environment
##########################
## Packages
library('pacman')
pacman::p_load(dplyr,rpart,FNN,performanceEstimation,unbalanced, caret, mgcv)
## User Defined Parameters
## KNN Objectives
Model_Evaluation <- 'DEPLOY' ## VALIDATE | TEST | DEPLOY
KNN_TrainSet_Change <- 'ENN' ## NONE | SCALE | SMOTE | TOMEK | ENN
## KNN Parameters
KNN_K = 19 ##Run once (K=1),then run 'TEST-K' script below for optimization
SMOTE_PrcOver = 3
SMOTE_PrcUnder = 1.34
ENN_K = 3
## Dataset Parameters
df_Size <- 10000 ## Magnitude of 10 (e.g., 10, 100, 1000, etc.)
df_Imbal <- 0.1 ## Works best <= 0.5
df_Bias <- 'BIAS' ## NONE | BIAS << None = Random, Bias = Bias data
## Splits
Pc_Train = .80
Pc_valid = .5 ##Allocates this percentage of remaining data to valid
## Data
set.seed(2023)
Index <- 1 + (0:(df_Size - 1)) * 1
df<-data.frame(IndexID=as.factor(Index),y=rep(as.factor(c('0','1'))
,times=c(ceiling(as.integer(df_Size*(1-df_Imbal)))
,ceiling(as.integer(df_Size*df_Imbal)))))
df$x1=if(df_Bias=='BIAS'){ifelse(df$y=='1',rnorm(sum(df$y=='1'),sd=0.5)
,rnorm(sum(df$y=='0')))}else{rnorm(df_Size)}
df$x2=if(df_Bias=='BIAS'){ifelse(df$y=='1',rnorm(sum(df$y=='1'),sd=0.5)
,rnorm(sum(df$y=='0')))}else{rnorm(df_Size)}
## Functions
knn_predict <-function(train_Data, test_Data, train_Class, K_param){
knn(train=train_Data,test=test_Data,cl=train_Class,prob=TRUE,k=K_param)}
knn_evaluate <- function(Type, Knn_Model, TestClass){
cm <- table ( TestClass, Knn_Model )
print ( cm )
if(all(c(0,1) %in% Knn_Model)) {
Accuracy <- round( (cm[1,1]+cm[2,2]) / sum(cm[]), digits = 4) * 100
Precision <- round( cm[2,2] / sum(cm[,2]), digits = 4)* 100
Specificity <- round( cm[1,1] / sum(cm[1,]), digits = 4)* 100
Recall <- round( cm[2,2] / sum(cm[2,]), digits = 4)* 100
F1Score<- round(2*((Precision*Recall)/(Precision+Recall)),digits=2)
print( paste('Model=',Type,': Accuracy=',Accuracy,'% | Precision='
,Precision,'% | Specificity=',Specificity,'% | Recall=',Recall
,'% | F1Score=',F1Score,'%'))
}else{print(paste('Model=',Model_Evaluation
,': Only one class predicted. Metrics cannot be calculated.'))}}
##########################
## KNN PREP: Split, Scale, SMOTE, TOMEK, ENN
##########################
## Shuffle & SPLIT
df <- sample_frac(df,1)
tIndex<-createDataPartition(df$y,p=Pc_Train,list=FALSE,times=1)
vIndex<-createDataPartition(df$y[-tIndex],p=Pc_valid,list=FALSE,times=1)
train <- df[tIndex,]
valid <- df[-tIndex,][vIndex,]
test <- df[-tIndex,][-vIndex,]
## SCALE
train_scale <- scale ( train[,3:4] )
test_scale <- scale ( test[,3:4] )
valid_scale <- scale ( valid[,3:4] )
## SMOTE
train_scale_smote <-
performanceEstimation::smote(y~x1+x2,data=mutate(as.data.frame(train_scale)
,y=train[,2]),perc.over=SMOTE_PrcOver,perc.under=SMOTE_PrcUnder)
## TOMEK
Tomek <- ubTomek( train_scale_smote[,-3], train_scale_smote[,3] )
train_scale_smote_TOMEK <- cbind(Tomek$Y, Tomek$X)
colnames( train_scale_smote_TOMEK) <- c("y", "x1" , "x2")
## ENN
ENN <- ubENN( train_scale_smote[,-3], train_scale_smote[,3] , k = ENN_K )
train_scale_smote_ENN <- cbind(ENN$Y, ENN$X)
colnames( train_scale_smote_ENN) <- c("y", "x1" , "x2")
##########################
## KNN
##########################
## KNN Evaluation Parameters
KNN_Train <-
if (KNN_TrainSet_Change %in% 'NONE') { train[,3:4]
} else if(KNN_TrainSet_Change %in% 'SCALE') {train_scale
} else if(KNN_TrainSet_Change %in% 'SMOTE') {train_scale_smote[,-3]
} else if(KNN_TrainSet_Change %in% 'TOMEK') {train_scale_smote_TOMEK[,-1]
} else if(KNN_TrainSet_Change %in% 'ENN') {train_scale_smote_ENN[,-1]
}
KNN_TrainClass <-
if (KNN_TrainSet_Change %in% c('NONE','SCALE')) {train[,2]
} else if (KNN_TrainSet_Change %in% 'SMOTE') {train_scale_smote[,3]
} else if (KNN_TrainSet_Change %in% 'TOMEK') {train_scale_smote_TOMEK[,1]
} else if (KNN_TrainSet_Change %in% 'ENN') {train_scale_smote_ENN[,1]
}
KNN_Test_Tr<-if(KNN_TrainSet_Change == 'NONE'){train[,3:4]}else{train_scale}
KNN_Test_V<-if(KNN_TrainSet_Change == 'NONE'){test[,3:4]}else{valid_scale}
KNN_Test_Tt<-if(KNN_TrainSet_Change == 'NONE'){valid[,3:4]}else{test_scale}
KNN_TestClass_Tr <- train[,2]
KNN_TestClass_V <- valid[,2]
KNN_TestClass_Tt <-test[,2]
## KNN MODELS
KNN_Model_Train <- knn_predict(KNN_Train, KNN_Test_Tr, KNN_TrainClass, KNN_K)
KNN_Model_Valid <- knn_predict(KNN_Train, KNN_Test_V, KNN_TrainClass, KNN_K)
KNN_Model_Test <- knn_predict(KNN_Train, KNN_Test_Tt, KNN_TrainClass, KNN_K)
## KNN as Feature Engine (PSEUDO-LABELING|SELF-TRAINING)
if(Model_Evaluation=='VALIDATE'){
knn_evaluate('Validate', KNN_Model_Valid, KNN_TestClass_V )} else
if(Model_Evaluation=='TEST')
{knn_evaluate('Test',KNN_Model_Test,KNN_TestClass_Tt)} else
if(Model_Evaluation=='DEPLOY') {
## Feature Engineering
## Training set for Ensemble Learning (Part 2)
KNN_Model_Final <- knn_predict(KNN_Train, KNN_Train,KNN_TrainClass, KNN_K)
KNN_Train_Final <- mutate(as.data.frame(KNN_Train),y=KNN_TrainClass)
KNN_Train_Final$Class <- KNN_Model_Final
KNN_Train_Final$ClassScore <- attr( KNN_Model_Final, "prob" )
##View Train Evaluation:
# knn_evaluate('FINAL', KNN_Train_Final[,4], KNN_Train_Final[,3] )
## Test & Validation sets for Ensemble (Part 2)
KNN_Test_Tr <- mutate(as.data.frame(KNN_Test_Tr),y=KNN_TestClass_Tr)
KNN_Test_Tr$Class <- KNN_Model_Train
KNN_Test_Tr$ClassScore <- attr(KNN_Model_Train, "prob")
KNN_Test_V <- mutate(as.data.frame(KNN_Test_V),y=KNN_TestClass_V)
KNN_Test_V$Class <- KNN_Model_Valid
KNN_Test_V$ClassScore <- attr(KNN_Model_Valid, "prob")
KNN_Test_Tt <- mutate(as.data.frame(KNN_Test_Tt),y=KNN_TestClass_Tt)
KNN_Test_Tt$Class <- KNN_Model_Test
KNN_Test_Tt$ClassScore <- attr(KNN_Model_Test, "prob" )
## Review Results So Far
df_results <- rbind ( KNN_Test_V, KNN_Test_Tr , KNN_Test_Tt )
knn_evaluate('DEPLOY',df_results[,4],df_results[,3])
##Clear Memory
rm(df_results)
}Code: Testing K (Part One Updated) (R)
k_values <- c(3, 5, 7, 15, 19) # List of K values to iterate over
for (k in k_values) {
KNN_Model_Valid <- knn_predict(KNN_Train, KNN_Test_V, KNN_TrainClass, k)
knn_evaluate(paste('K=', k, sep=''), KNN_Model_Valid, KNN_TestClass_V)
}Code: Ensemble Learning (Part Two)
##########################
## Establish Environment
##########################
## User Defined Parameters
Model_Evaluation <- 'VALIDATE' ## VALIDATE | TEST | DEPLOY
FeatureEngine_Avail <- 'N' ## Y | N
Tree_y_Weight = .5 ## 0 through 1
Tree_cp = .01
Tree_minsplit = 10
## Other Parameters
set.seed(2023)
## Functions
Model_Predict<-function(model,eval_data,model_name){
predictions <- if(model_name=='Tree'){predict(model,newdata=eval_data)}else
predict(model, newdata = eval_data, type = "response")
ifelse(predictions >= .5, 1, 0)
}
Model_Evalaute<-function(model,eval_data_class,model_name){
cm <- if (model_name == 'Tree') {
table ( eval_data_class, model[,2] )
} else table ( eval_data_class, as.vector(model) )
print ( cm )
if(all(c(0,1) %in% model)) {
Accuracy <- round( (cm[1,1]+cm[2,2]) / sum(cm[]), digits = 4) * 100
Precision <- round( cm[2,2] / sum(cm[,2]), digits = 4)* 100
Specificity <- round( cm[1,1] / sum(cm[1,]), digits = 4)* 100
Recall <- round( cm[2,2] / sum(cm[2,]), digits = 4)* 100
F1Score<- round(2*((Precision*Recall)/(Precision+Recall)),digits=2)
print( paste('Model=',model_name,': Accuracy=',Accuracy,'% | Precision='
,Precision,'% | Specificity=',Specificity,'% | Recall=',Recall
,'% | F1Score=',F1Score,'%'))
}else{print(paste('Model=',model_name
,': Only one class predicted. Metrics cannot be calculated.'))}
}
## Other
if(FeatureEngine_Avail=="Y"){
Columns<-c('y','x1','x2','Class')#,'ClassScore')
selected_cols<-c('x1','x2','Class')#,'ClassScore')
Formula=y~x1+x2+Class#+ClassScore
} else if (FeatureEngine_Avail=="N"){
Columns<-c('y','x1','x2')
selected_cols<-c('x1','x2')
Formula=y~x1+x2
}
##########################
## ENSEMBLE PREP
##########################
Enmbl_Training <- KNN_Train_Final[Columns]
Enmbl_Vld <- KNN_Test_V[Columns] ## Ensemble Valid
Enmbl_Tst <- KNN_Test_Tt[Columns] ## Ensemble Test
Enmbl_Trn <- KNN_Test_Tr[Columns] ## Ensemble Train
weights = ifelse(Enmbl_Training$y == 1, Tree_y_Weight, (1-Tree_y_Weight))
##########################
## ENSEMBLE MODELS
##########################
## Models || GAM note: variable optimization: y ~ s(x)...
GAM<-gam(as.formula(Formula),data=Enmbl_Training,family=binomial())
GLM<-glm(y~.,data=Enmbl_Training,family=binomial(link="logit"))
Tree<-rpart(y~.,data=Enmbl_Training,method="class", weights = weights
,control = rpart.control(cp=Tree_cp, minsplit = Tree_minsplit)
)
## Predictions
GAM_Pred_Tr<-Model_Predict(GAM,as.data.frame(Enmbl_Trn[selected_cols]),'GAM')
GAM_Pred_V<-Model_Predict(GAM,as.data.frame(Enmbl_Vld[selected_cols]),'GAM')
GAM_Pred_Tt<-Model_Predict(GAM,as.data.frame(Enmbl_Tst[selected_cols]),'GAM')
GLM_Pred_Tr<-Model_Predict(GLM,as.data.frame(Enmbl_Trn[selected_cols]),'GLM')
GLM_Pred_V<-Model_Predict(GLM,as.data.frame(Enmbl_Vld[selected_cols]),'GLM')
GLM_Pred_Tt<-Model_Predict(GLM,as.data.frame(Enmbl_Tst[selected_cols]),'GLM')
Tree_Pred_Tr<-Model_Predict(Tree,as.data.frame(Enmbl_Trn[selected_cols]),'Tree')
Tree_Pred_V<-Model_Predict(Tree,as.data.frame(Enmbl_Vld[selected_cols]),'Tree')
Tree_Pred_Tt<-Model_Predict(Tree,as.data.frame(Enmbl_Tst[selected_cols]),'Tree')
## Evaluations
if (Model_Evaluation == "VALIDATE") {
Model_Evalaute(GAM_Pred_V,Enmbl_Vld$y,'GAM' )
Model_Evalaute(GLM_Pred_V,Enmbl_Vld$y,'GLM' )
Model_Evalaute(Tree_Pred_V,Enmbl_Vld$y,'Tree' )
} else if (Model_Evaluation == "TEST") {
Model_Evalaute(GAM_Pred_Tt,Enmbl_Tst$y,'GAM' )
Model_Evalaute(GLM_Pred_Tt,Enmbl_Tst$y,'GLM' )
Model_Evalaute(Tree_Pred_Tt,Enmbl_Tst$y,'Tree' )
} else if(Model_Evaluation=='DEPLOY') {
## Predict Over Train
Enmbl_Trn$GAMClass <- GAM_Pred_Tr
Enmbl_Trn$GLMClass <- GLM_Pred_Tr
Enmbl_Trn$TreeClass <- Tree_Pred_Tr[,2]
## Predict Over Valid
Enmbl_Vld$GAMClass <- GAM_Pred_V
Enmbl_Vld$GLMClass <- GLM_Pred_V
Enmbl_Vld$TreeClass <- Tree_Pred_V[,2]
## Predict Over Test
Enmbl_Tst$GAMClass <- GAM_Pred_Tt
Enmbl_Tst$GLMClass <- GLM_Pred_Tt
Enmbl_Tst$TreeClass <- Tree_Pred_Tt[,2]
## Combine
Ensemble_Final<-rbind(Enmbl_Trn,Enmbl_Vld,Enmbl_Tst)
## Evaluate >> Code differently based on variables available
if ( FeatureEngine_Avail == 'Y' ) {
Model_Evalaute (Ensemble_Final[,5],Ensemble_Final[,1],'GAM' )
Model_Evalaute (Ensemble_Final[,6],Ensemble_Final[,1],'GLM' )
Model_Evalaute (Ensemble_Final[,7],Ensemble_Final[,1],' Tree' )
} else {
Model_Evalaute (Ensemble_Final[,4],Ensemble_Final[,1],'GAM' )
Model_Evalaute (Ensemble_Final[,5],Ensemble_Final[,1],'GLM' )
Model_Evalaute (Ensemble_Final[,6],Ensemble_Final[,1],' Tree' )
}
}Part Two
Overview
This section will evaluate the impact of using K-Nearest Neighbors (KNN) as a feature engine on our imbalanced dataset, where the data distribution is skewed towards one class. The output of the KNN algorithm will serve as an attribute for subsequent ensemble learning models, a combination of various machine learning algorithms to achieve better predictive performance. However, it is important to note that our findings here may not necessarily guarantee similar results in different contexts. Our primary aim is to present a comprehensive example, highlight significant methodologies, and provide you with an additional tool for your data science toolkit.
With that said, let’s dive right into the code!
Section One: Establish Environments
Section one is broken into two main subsections: User Defined parameters and ‘Other’. We will cover each subsection respectively.
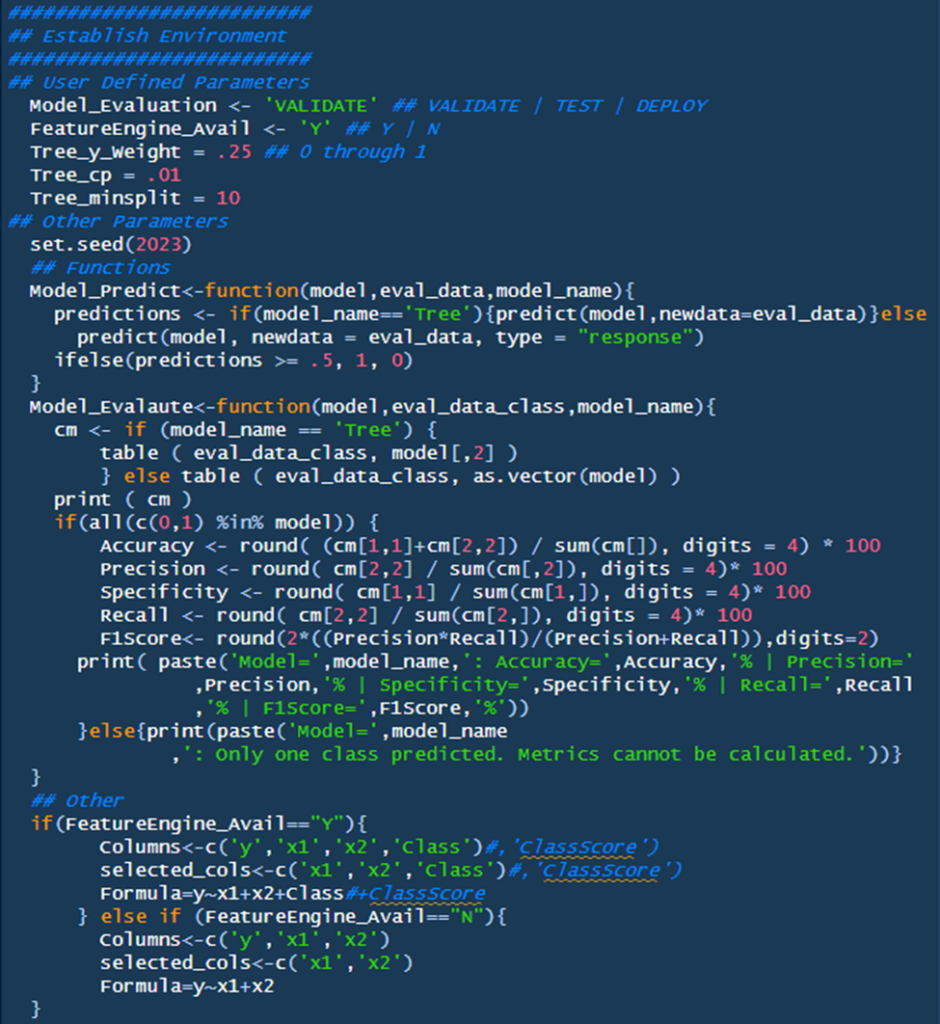
- User Defined Parameters
- Model_Evaluation: This determines the model’s evaluation. Validate runs the model over the ‘validation’ dataset, ‘test’ runs the model over the test dataset, and ‘deploy’ evaluates the whole model generating an output.
- FeatureEngine_Avail: This includes/excludes the KNN-Feature-Engine parameters from part one. Y includes, N excludes.
- Tree_y_Weight: Controls the weights parameter from rpart. Technical information on the parameter can be found here (Cran, rpart), a general overview can be found here (StackOverlow, “How to apply weights in rpart?”)
- Tree_cp: Controls the cp parameter from rpart; it’s a pruning technique that functions as follows: “don’t split a partition if the new partition does not ‘significantly’ reduce the impurity” (Bruce & Bruce, p.226). Technical information on the parameter can be found here (Cran, rpart), a general overview can be found here (Analytics Vidhya, “How does Complexity Parameter (CP) work in decision tree”
- Tree_minsplit: Controls the minsplit parameter from rpart. It’s a pruning technique where, “Avoid splitting a partition if a resulting sub partition is too small, or if a terminal leaf is too small” (Bruce & Bruce, p.226). Technical information on the parameter can be found here (Cran, rpart), a general overview can be found here (Medium: “Minsplit and Minbucket”)
- Other Parameters
- Set.seed: enables replicable randomness. More information here (Statology, “How (And When) to Use set.seed in R”)
- Functions
- Model Predict: accepts inputs and outputs the ‘predictions’ from the desired model
- Input 1 – model: this is the model as defined in section ‘Ensemble Models’ > ‘Models’
- Input 2 – eval_data: this is the dataset to be predicted over
- Input 3 – model_name: this is a user defined model name. Please use ‘Tree’ for decision trees
- Model_Evaluate: accepts inputs and evaluates the output from Model_Predict
- Input 1 – model: this is the model to be evaluated from ‘Ensemble Models’ > ‘Predictions’
- Input 2 – eval_data_class: these are the respective classifiers for the Model_Predict eval_data
- Input 3 – model_name: this is a user defined model name. Please use ‘Tree’ for Decision trees
- Model Predict: accepts inputs and outputs the ‘predictions’ from the desired model
- Other: These are other dynamic parameters that update based on the output from “FeatureEngine_Avail”
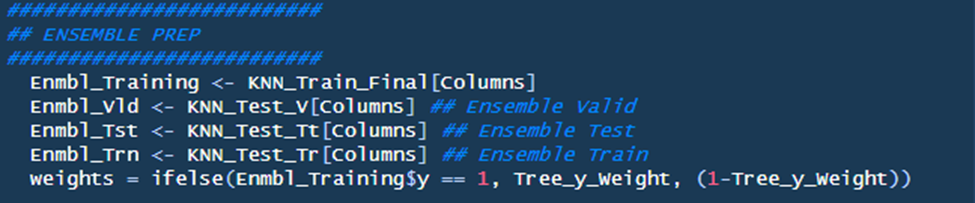
Section Two: Ensemble Prep
This is a very simply section that establishes ensemble dataset names. Additionally, weights are created based on the User Defined Parameter ‘Tree_y_Weight’ and the Enmbl_Training dataset.
Section Three: Ensemble Models
This section procures the models, makes predictions, and outputs the evaluations based on the user defined response for ‘Model_Evaluation’.
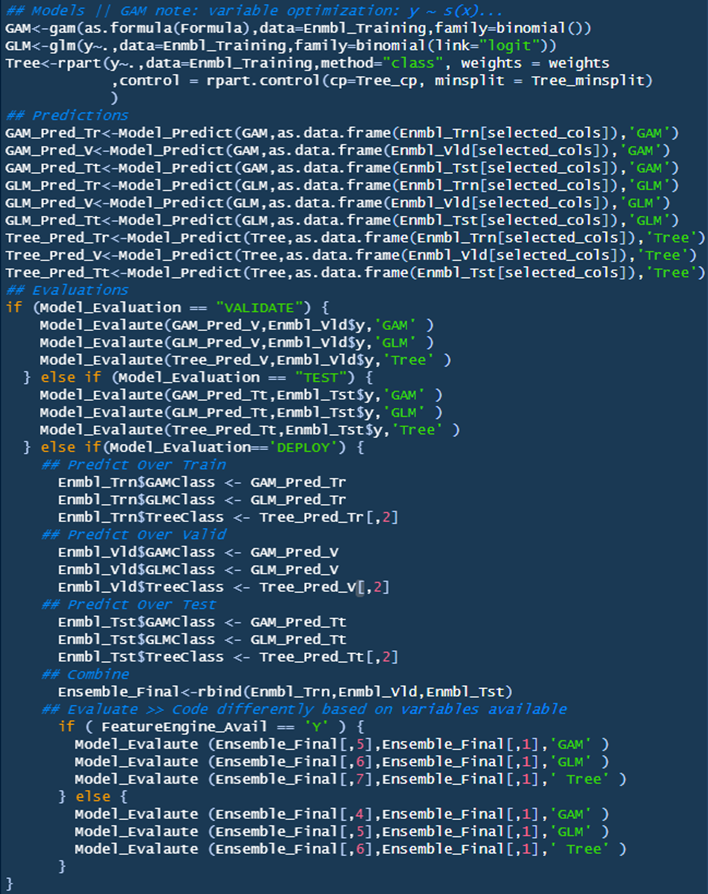
Models
Three models are being utilized in this ensemble script: GAM, GLM, and Decision Tree. Each will be evaluated separately to review their efficacy over this dataset. As noted in the script, GAM can be further optimized by adding automatic knots for spline terms (Bruce & Bruce, p.170); a good overview can be found here (Stackoverflow, “mgcv: How to set number and or locations of knots for splines”).
Predictions
Predictions are evaluated for each model over each dataset (train/valid/test). This is done so predictions aren’t duplicated based on the ‘model evaluation’ (i.e., reduce duplicative code). Terms for the custom function can be found above for the ‘Model_Evaluate’ function.
Evaluations
This section evaluates the model based on the user defined ‘Model_Evaluation’. Model_Evaluation == VALIDATE will predict and evaluate over the validate set, Model_Evaluation == TEST will predict and evaluate over the test set, and Model_Evalaution == DEPLOY will predict over the train, validate, and testing sets, merge the results, and then procure the results based on if the user added the KNN_FeatureEngine.
Results
Let me reiterate that we have developed three models which offer users a variety of training options for their datasets. It’s crucial to remember that these models’ performance may vary across different applications, so the superior performance of a model on a randomized dataset does not automatically translate into all scenarios. Now, let’s explore the performance of these models!
Model Comparison
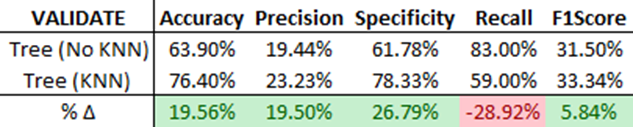
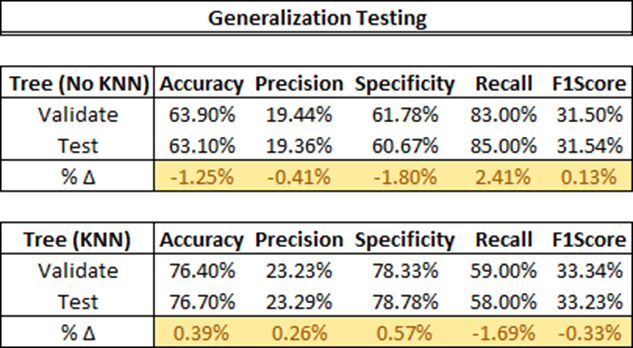
Initially, we test our ensemble method’s efficacy by comparing results with and without the KNN Feature Engine. Without KNN features, the highest performing ‘Validate’ model was the Decision Tree Model (weight of 0.5). With KNN features, it was the same Decision Tree Model, but with a reduced weight of 0.25. The comparison of the test outputs is shown below.
Model Evaluation on New Data
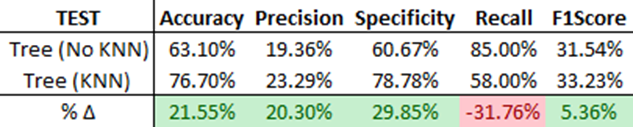
Next, we evaluate each model on new data (‘TEST’) and assess their generalization capabilities. The Decision Tree Model with KNN once again outshines the same model without KNN, showing improvement in all performance metrics except Recall. This can be explained by the precision-recall trade-off, a phenomenon where an increase in precision often leads to a decrease in recall (For more information on the precision-recall trade-off, refer to this [Medium article]). However, this relationship isn’t always the case. Both models generalize well with a deviance of less than 5%.
Model Selection and Deployment
Finally, we select and deploy the highest-performing model, the Decision Tree with KNN features, and compare its performance against random chance and the standalone KNN model.
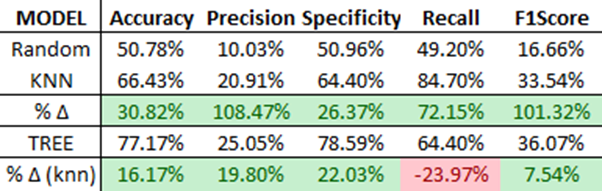
As shown, the ensemble Decision Tree model outperforms the standalone KNN model in all scenarios except recall. Interestingly, without the KNN feature engine, the ensemble model wouldn’t perform as well as the standalone KNN model! You can observe this by evaluating the earlier ‘Tree (No KNN)’ model.
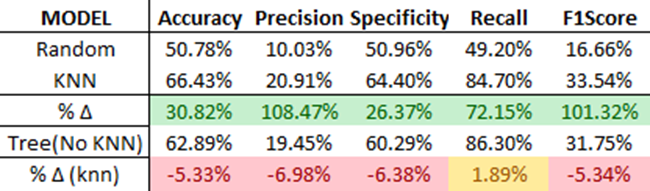
Conclusion
From these results, it’s fair to say that incorporating KNN as a feature engine significantly enhances the performance of the Decision Tree Model.
Final Consideration
In conclusion, this script is intended to enrich your data science toolkit. For future enhancements, we suggest incorporating auto-hyperparameter tuning and delving deeper into each model’s optimization. Even in its current form, we believe this script and its analysis can effectively guide users on how to utilize KNN as a feature engine for imbalanced datasets. We hope you find this resource valuable and look forward to your feedback.
Addendum: Part 1 Updates
Code Overview
Below are sections with significant code differences. These are quick explanations.
Section: Establish Environments
- User Defined Functions
- (Splits) Pc_Train is the percentage of the data frame used for training data.
- (Splits) Pc_Valid is the percentage of the remaining dataset used for the valid dataset
- Functions
- knn_predict: This function evaluates the KNN model over the inputs. train_Data is the training dataset, test_Data is the testing data dataset, train_Class is the classification data for the train_Data, and K_param is the K value for the model.
- Knn_evaluate: This function evaluates the KNN model from knn_predict. Type is a user defined model type (e.g., test, validate, etc.), Knn_Model is the KNN model to be evaluated, TestClass are the classifications for the test_Data used in the knn_predict model.
Section: KNN Prep
Shuffle & Split: This section now splits into three datasets for train, valid, and test
Section: KNN
This section now encompasses the old ‘KNN (Training)’ and ‘KNN as a Feature Engine’ sections.
KNN Models: Updated for three models for train, test, and validate.
KNN as a Feature Engine: This section now deploys the appropriate KNN model and evaluation based on the user defined Model_Evaluation. The final ‘Deploy’ section deploys predictions to the train, test, valid, and training dataset. The difference between train and training datasets is that the train dataset is pre-SMOTE data. The train, test, and valid sets are merged for evaluation and then cleared from memory for space.
Section: Testing K
This section greatly simplifies the KNN testing script as it now loops through custom functions based on the defined k_values provided. Update these k_values to update k-tested values.
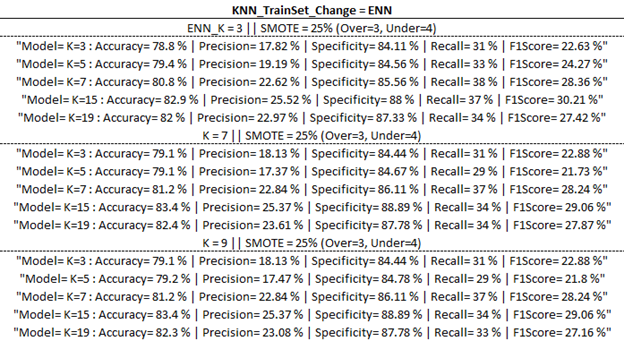
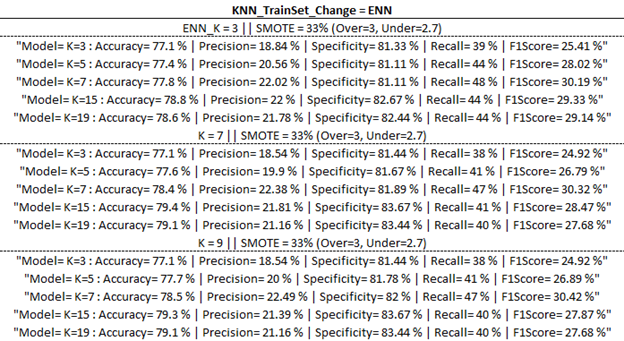
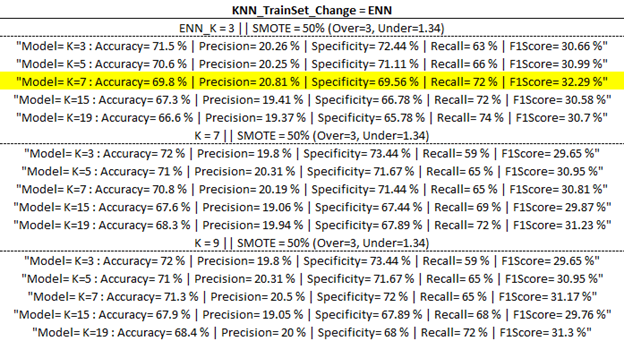
Training Model Outputs
Below are the training model outputs with the highest F1 score per category (corresponding to ‘Best Output’ per ‘part one updates’ table above) highlighted in yellow.
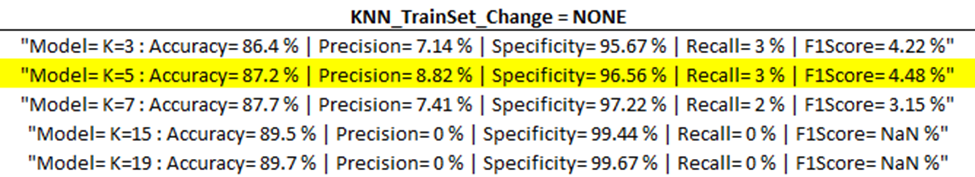
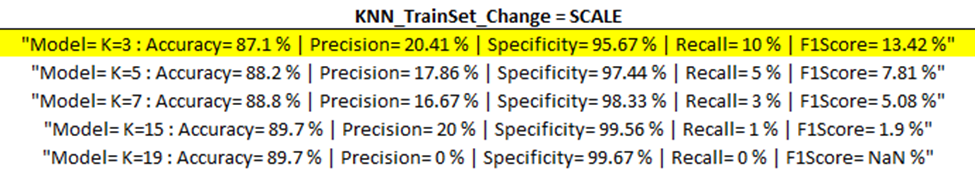
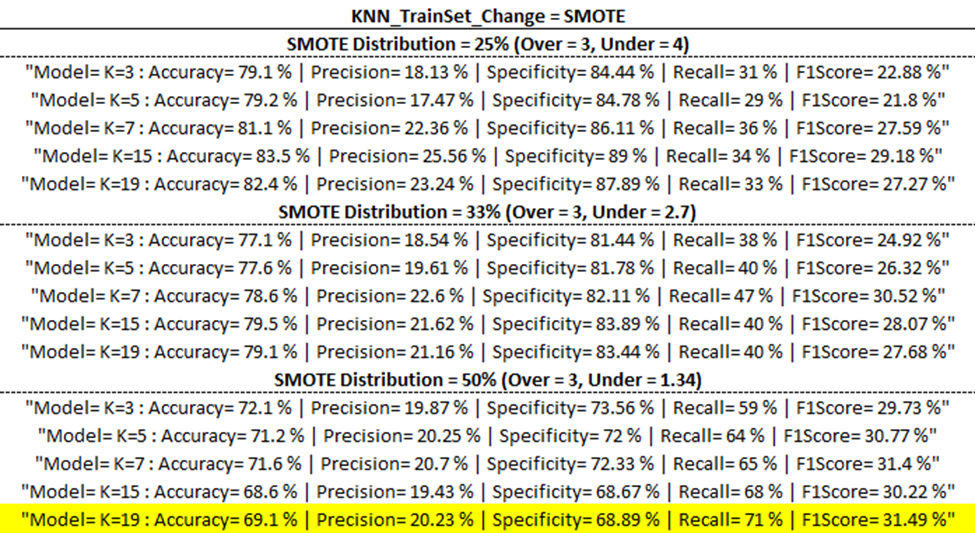
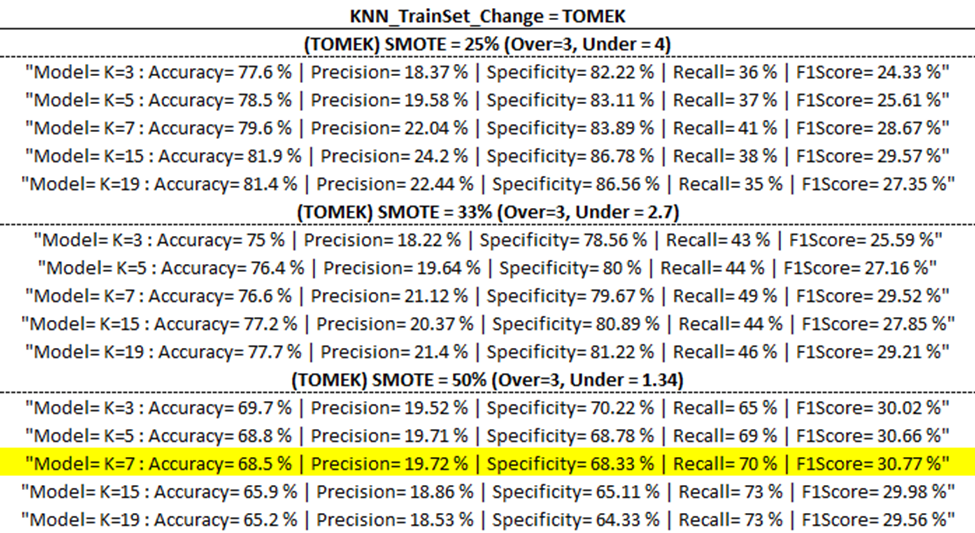
(ENN)
REFERENCES
“Practical Statistics for Data Scientists”, Peter Bruce & Andrew Bruce, (Amazon Affiliate Link) https://amzn.to/3OIQe5z

